Laboratoire
Ancestral role of DNA mechanics in bacterial transcriptional regulation
A paper by the CRP team in Nucleic Acids Research: Quantitative contribution of the spacer length in the supercoiling-sensitivity of bacterial promoters
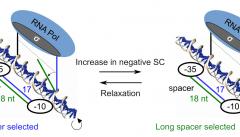
DNA supercoiling acts as a global transcriptional regulator in bacteria, but the promoter sequence or structural determinants controlling its effect remain unclear. It was previously proposed to modulate the torsional angle between the -10 and -35 hexamers, and thereby regulate the formation of the closed-complex depending on the length of the “spacer” between them. Here, we develop a thermodynamic model of this notion based on DNA elasticity, providing quantitative and parameter-free predictions of the relative activation of promoters containing a short vs long spacer when the DNA supercoiling level is varied. The model is tested through an analysis of in vitro and in vivo expression assays of mutant promoters with variable spacer lengths, confirming its accuracy for spacers ranging from 15 to 19 nucleotides, except those of 16 nucleotides where other regulatory mechanisms likely overcome the effect of this specific step. An analysis at the whole-genome scale in E. coli then demonstrates a significant effect of the spacer length on the genomic expression after transient or inheritable superhelical variations, validating the model’s predictions. Altogether, this study shows an example of mechanical constraints associated to promoter binding by RNA Polymerase underpinning a basal and global regulatory mechanism.
This paper has been slected by CNRS to illustrate its monthly science facts & news: https://www.insb.cnrs.fr/fr/cnrsinfo/la-torsion-mecanique-de-ladn-aide-larn-polymerase-se-fixer-aux-promoteurs-bacteriens
Bravo to Sam and all authors (for the science and for this specific achievement :-).

