Regulation of pectinase gene expression
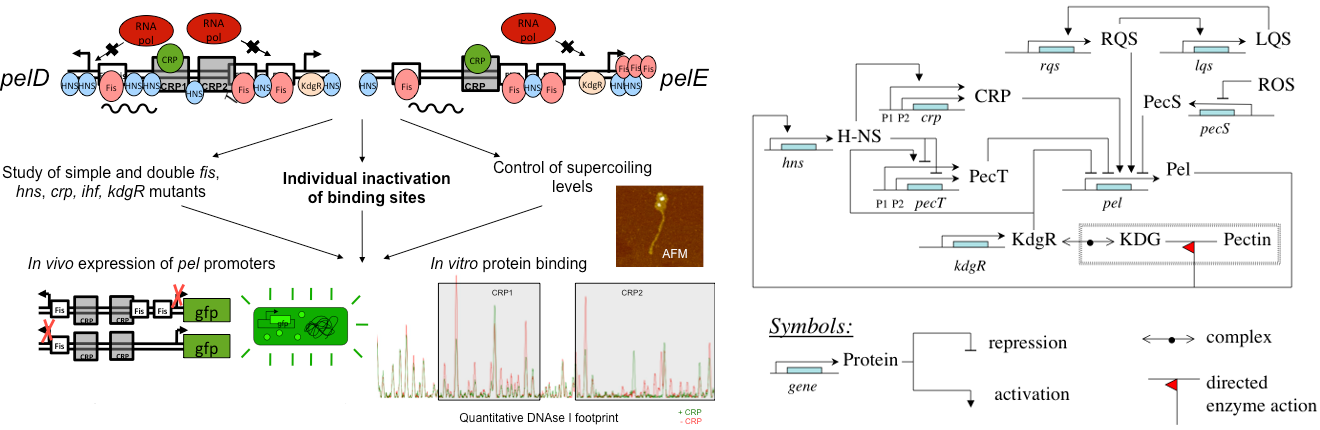
Our first research axis is devoted to the characterization of nucleoprotein complexes assembly at the promoters of pectinase genes, quantitative modelling of their expression as well as their evolution. The dual function of pectate lyases in virulence and nutrition implies that expression of pel genes is regulated by both metabolic and virulence regulators. Dickeya senses pectin via the specific repressor KdgR and couples pel gene expression to central metabolism via the global activator CRP. KdgR represses the whole pectin catabolic pathway in the absence of the corresponding substrate whereas CRP ensures the preferential utilization of glucose or other easily metabolizable carbohydrates, when bacteria are in the presence of a mixture of carbon sources. Regarding the different expression patterns of the two major virulence pelD and pelE paralogs (Duprey et al. 2016), we wish to develop a quantitative modelling for these two genes incorporating the impact of metabolites (KDG and cAMP, which modulate the activities of the regulators KdgR and CRP), DNA supercoiling and NAPs (FIS, H-NS, IHF, lrp). In addition, we will analyse the structural and elastic properties of DNA sequences involved in indirect sequence readout by regulators such as PecS, PecT, VfmE. Modelling of these interplays will predict the dynamics of nucleoprotein complex assembly during infection and the resulting expression patterns.
Selected bibliography:
- Jacques-Alexandre Sepulchre, Sylvie Reverchon, Jean-Luc Gouzé, William Nasser. Modeling the bioconversion of polysaccharides in a continuous reactor: A case study of the production of oligo-galacturonates by Dickeya dadantii. Journal of Biological Chemistry, American Society for Biochemistry and Molecular Biology, 2019, 294 (5), pp.1753-1762. 〈10.1074/jbc.RA118.004615〉. 〈hal-01952846)
- O. Zghidi-Abouzid, E. Herault, S. Rimsky, S. Reverchon, W. Nasser, et al.. Regulation of pel genes, major virulence factors in the plant pathogen bacterium Dickeya dadantii, is mediated by cooperative binding of the nucleoid-associated protein H-NS. Research in Microbiology, Elsevier, 2016, 167 (4), pp.247-253. 〈hal-01997084〉
- A. Duprey, G. Muskhelishvili, S. Reverchon, W. Nasser. Temporal control of Dickeya dadantii main virulence gene expression by growth phase-dependent alteration of regulatory nucleoprotein complexes. Biochimica et Biophysica Acta - Gene Regulatory Mechanisms , Elsevier, 2016, 1859 (11), pp.1470-1480. 〈hal-01997030〉