Chromatin dynamics and gene expression
One major objective of the CRP team is to characterize the relation between chromosome configuration and genome expression at the global scale, based on the phytopathogenic bacterium Dickeya dadantii. This "analogue" mode of regulation, currently underestimated and almost absent of modeling works, differs from classical "digital" regulation, by the involved molecular actors (Nucleoid Associated Proteins and DNA supercoiling vs transcription factors) as well as by its global action on the genome. Methods used by the team are transcriptomics, chromosome manipulation, and computational and biophysical modeling.
1. Characterization of analogue and digital contributions in transcriptional regulation
To understand the regulatory network responsible for the expression of virulence genes and therefore the success of infection, we use novel approaches for analyses of global gene expression that establish correlations between the physical-dynamical properties of DNA sequences of expressed genes (such as melting energy, sensitivity to DNA relaxation) and DNA structuring effects of Nucleoid Associated Proteins. Using this systemic approach, we demonstrated, in D. dadantii, the existence of chromosomal domains in which numerous genes are either activated or repressed in a coherent manner in response to various environmental stresses, likely in relation with the three-dimensional folding of the chromosome, whereas their differential response is linked to physicochemical properties of encoding DNA sequences. Comparing our data with in planta expression patterns reveals the combined impact of stressors and allows us to predict what stress D. dadantii is experiencing during the different stages of infection (Jiang et al. 2015 and 2016). In collaboration with M. Hütt at Jacobs Univ. (Bremen, Germany) and G. Muskhelishvili (Tbilissi, Georgia), we are now developing an analysis of transcriptomics data, with the aim of separating the analogue and digital contributions in conditions relevant to the infection process and with NAP mutant strains (Meyer et al. 2018, Muskhelishvili et al. 2019).
2. Large-scale chromosome dynamics and transcriptional activity
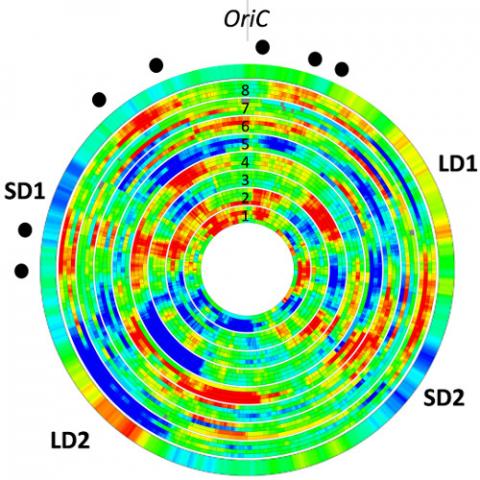
Previous analyses have demonstrated the existence of transient large-scale chromosomal domains of coherent gene expression in Dickeya (Jiang et al. 2015). We address now the attractive hypothesis that, at this scale, the chromosome dynamics is coupled to its transcriptional activity (possibly in both directions), which can be physically monitored. To test and quantify this effect, we have developed a collaboration with the team of Christian Lesterlin (IBCP, Lyon) in super-resolution imaging of fluorescent tags within live cells. The projects consists in following the localisation of different chromosomal loci as well as NAPs in different environmental conditions where domains are activated or repressed, and using different mutant strains (i.e., with displaced NAP genes). The working hypothesis is that the spatial organisation of the chromosome is related to its functional state (expressed regions being typically shifted outwards in the cell), and the objective is to decipher the specific role of NAPs in this mechanism.
3. Global transcriptional regulation by DNA supercoiling
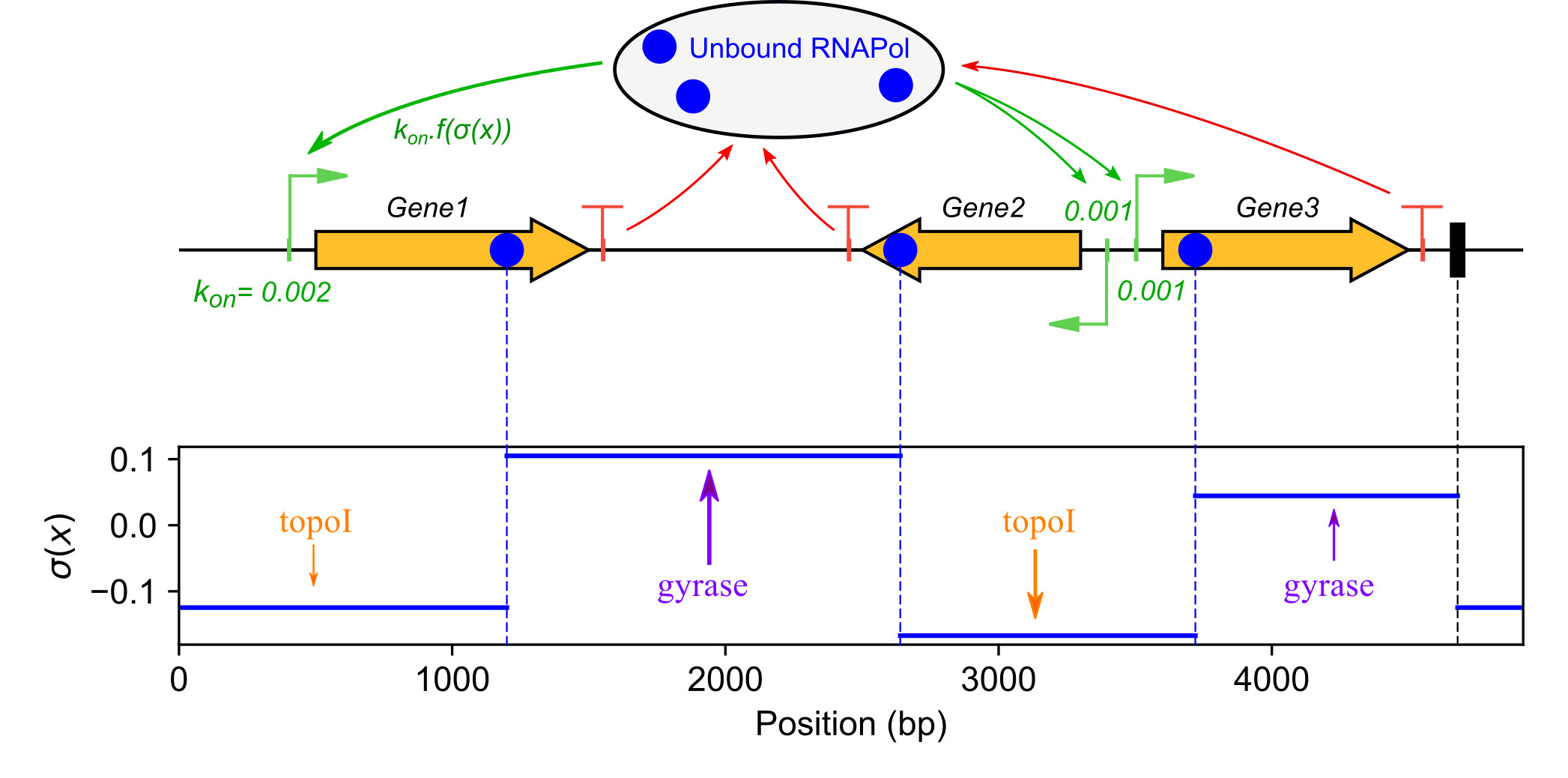
We develop quantitative models of the global regulation of bacterial transcription by DNA mechanics, and more specifically DNA supercoiling (Martis et al. 2019). The effect of the latter on genes' activity is explained according to (1) their promoter structure and (2) spatial supercoiling heterogeneities along the chromosome. Transcription initiation is described by thermodynamic models of the DNA-RNA Polymerase interaction, exhibiting the role (i) the spacer length between the -10 and -35 binding sites of the RNA Polymerase affecting the formation of the closed-complex, and (ii) the discriminator sequence (between the -10 box and the TSS) affecting DNA opening required for open-complex formation. These two structural elements are unbound by any regulatory protein and may yet explain why different genes respond differently to global superhelical variations in the chromosome. This hypothesis is tested using published and original transcriptomics data from evolutionarily distant species, and more specific experiments in Dickeya. The spatial variations of DNA supercoiling along the Dickeya chromosome are analysed using new data obtained with the intercalating agent tri-methyl-psoralen, in collaboration with Monika Glinkowska (Univ. Gdansk, Poland). This analysis is complemented with modeling work (El Houdaigui et al. 2019), and this coupled effort is funded by an ANR JCJC grant (LoToReg 2018).