Enoch Yeung: Exploring DNA Twisting as a Hypothesis to Explain Variation in mRNA Expression in the Genome (Séminaire Externe MAP - Equipe CRP)
Dr Enoch Yeung de l'université UCSB de Santa Barbara, invité par l'Equipe CRP
Abstract: DNA supercoiling is a biophysical mechanism for controlling gene transcription in bacteria. Integrating new genes can disrupt local supercoiling in genomic locus, altering expression of neighboring endogenous genes. Despite having established methods for genomic integration, genomic integration sites (or landing sites) are often chosen without predictive modeling of biophysical impacts. This work takes steps towards a rigorous methodology to design insulated genomic landing sites in bacteria, using a combination of targeted genomic engineering and biophysical modeling. First, we examine the effect of insertion sequence content and length at a select set of loci in E. coli. We select sites that meet standard criteria for integration, namely separation from essential genes and adequate separation from transcriptionally active, neighboring genes. Second, we conduct a detailed, biophysical case study on one such integration locus. We show the expression levels of reporter genes as well as a list of essential genes in E. coli respond to changes in relative orientation of genes within the integration site.
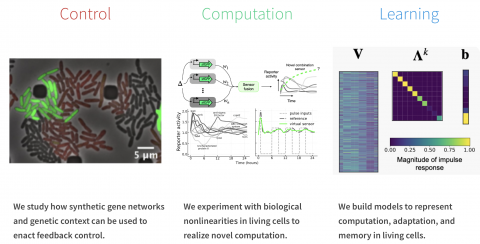
Biosketch: Enoch Yeung is an Associate Professor in the Department of Mechanical Engineering at the University of California, Santa Barbara (UCSB). Prior to his appointment at UCSB, Enoch was a Senior Research Scientist for the Pacific Northwest National Laboratory (PNNL), working on research problems at the interface of synthetic biology and machine learning, harnessing the genetics of non-model microbes, and new artificial intelligence algorithms to bootstrap synthetic biology design and analysis in non-model microbes. At UCSB, he is studying how genetics and dynamics of gene expression in microbes can be blended and harnessed to enact new biophysical mechanisms for transcriptional control and biosensing. Recently, he demonstrated an end-to-end experimental to analytics pipeline for detecting novel biomarkers responsive to novel chemical compounds. He earned his PhD from the California Institute of Technology in 2016 and his Bachelors in 2010 from BYU with University Honors, magna cum laude. He is a recipient of the National Science Foundation CAREER award, the Army Young Investigator Program award, and the Keck Foundation award, and the Pacific Northwest National Laboratory Outstanding Performance Award. More broadly, his research interests are in biological control, biological sensing, DNA biophysics, robust biological design, and quantitative methods for accelerating biological analysis.

Informations complémentaires
- sam.meyer@insa-lyon.fr
-
salle McClintock, bâtiment Pasteur, INSA Lyon